In the intricate ballet of cellular processes, precise regulation of the cell cycle is crucial for healthy cell function and reproduction. A new study, led by researchers Jordan Xiao, Jonathan J Turner, Mardo Kõivomägi, and Jan M Skotheim, delves into the microscopic intricacies governing cell cycle dynamics, focusing on the phosphorylation of the transcriptional inhibitor Whi5 in budding yeast. This protein, pivotal for the transition from G1 phase to S phase, undergoes critical changes in phosphorylation that decide when a cell will commit to DNA replication and subsequent division.
At the heart of this study, published in the journal [Journal Name], is the exploration of how different phosphorylation states of Whi5—hypo-phosphorylation in early G1 and hyper-phosphorylation in late G1—impact its role and functionality. Through meticulous experimentation, the team identified 19 phosphorylation sites on Whi5, pinpointing seven critical for maintaining its hypo-phosphorylated state during early G1. Alterations in these sites were found to affect cell size and delay the crucial G1/S transition.
Moreover, the research highlights the role of “priming” phosphorylation sites necessary for rapid hyper-phosphorylation later in G1, which facilitates Whi5’s exit from the nucleus, enabling the progression to subsequent cell cycle phases. This groundbreaking study not only refines the understanding of Whi5’s role in G1/S transition but also extends its importance to the entire cell cycle, offering potential new avenues for understanding cell cycle regulation in more complex organisms.
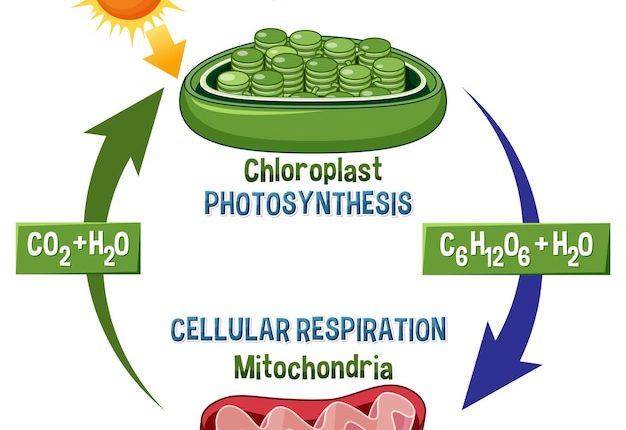
Understanding the molecular underpinnings of cell cycle regulation is a central quest in cell biology. Fundamental to this regulation is the proper progression through the cell cycle, a series of phases allowing cells to grow and duplicate their DNA before dividing. The transition between these phases must be exquisitely controlled to maintain cellular health and function. Mishaps in this regulation often result in uncontrolled cell growth, a hallmark of cancer, making this field not only fascinating but also crucial for its implications in health and disease.
The transcriptional inhibitor Whi5 in budding yeast (Saccharomyces cerevisiae) plays a vital role similar to the retinoblastoma (Rb) protein in mammals. It acts as a gatekeeper for the G1/S transition in the cell cycle, making its investigation particularly relevant for understanding similar mechanisms in human cells. During the G1 phase, cells prepare for DNA replication and subsequent division. This involves crucial decision-making processes dictated by internal and external cues that ensure the cell is ready to proceed to the next phase.
Research into Whi5 has revealed that its regulatory role is controlled by phosphorylation, a common form of protein modification that can alter a protein’s function, typically turning it on or off. Phosphorylation of Whi5 dictates its interaction with the transcription machinery, and thus its ability to control the expression of genes required for S phase entry. Early in G1 phase, Whi5 is less phosphorylated (hypo-phosphorylated) and binds to DNA to inhibit the expression of genes necessary for DNA replication. As cells approach the end of G1, a wave of phosphorylation (hyper-phosphorylation) displaces Whi5 from DNA, lifting the inhibition and allowing the cell to progress to the S phase.
The study led by researchers Jordan Xiao, Jonathan J Turner, Mardo Kõivomägi, and Jan M Skotheim provides deeper insights into this regulatory mechanism. By examining the specific phosphorylation sites on Whi5, their research not only illuminates how modifications at these sites influence its activity but also sheds light on the timing and sequence of phosphorylation events during G1. Understanding these dynamics is crucial for grasping how cells integrate multiple signals to make pivotal decisions about growth and division.
This detailed examination unfolds against a broader backdrop of studies focusing on cell cycle dynamics. Previous research has predominantly centered on how external factors such as nutrients and mating pheromones influence cell cycle progression in yeast. The efficacy of these extrinsic signals must ultimately be funneled through intrinsic regulators like Whi5, which integrate and execute the cell’s response. This integration ensures that cells only commit to DNA replication and subsequent division when conditions are optimal, thereby safeguarding genetic integrity and cell viability.
By elucidating the nuanced roles of phosphorylation states of Whi5, this study not only augments our understanding of cell cycle regulation in yeast but also serves as a model for similar mechanisms in higher organisms, including humans. Such insights offer potential avenues for therapeutic intervention in diseases characterized by dysregulated cell growth, such as cancer.
The research team employed a comprehensive suite of experimental techniques to dissect the phosphorylation dynamics of Whi5 and unravel its implications for cell cycle regulation in budding yeast.
**1. Phosphorylation Site Mapping and Mutagenesis:**
To identify the critical phosphorylation sites on Whi5, the team used mass spectrometry-based proteomics. This method allowed them to pinpoint the exact locations of phosphate groups added during the cell cycle phases. Following this extensive mapping, the researchers engineered yeast strains with specific mutations at these phosphorylation sites. By substituting serine or threonine (amino acids typically phosphorylated) with alanine, which cannot be phosphorylated, they created hypo-phosphorylatable mutants of Whi5. Conversely, substituting these residues with aspartic acid, mimicking a phosphorylated state, yielded hyper-phosphorylatable mutants. These genetically altered strains enabled the team to observe the effects of these mutations on cell cycle progression and Whi5 functionality.
**2. Fluorescence Microscopy and Time-Lapse Imaging:**
The cellular localization and timing of Whi5 activity were assessed using fluorescence microscopy. Whi5 was tagged with a fluorescent marker, allowing real-time visualization of its presence in the nucleus versus the cytoplasm. Time-lapse imaging techniques captured the dynamics of Whi5 during the G1 to S phase transition, providing insights into how changes in phosphorylation states influenced its intracellular movement.
**3. Flow Cytometry:**
To analyze cell cycle progression quantitatively, the researchers utilized flow cytometry. This technique measured DNA content across different cell cycle phases, helping to determine if mutations in Whi5 phosphorylation sites caused delays or advancements in the transition from G1 to S phase. Flow cytometry data corroborated findings from microscopy, offering a broader quantitative perspective on cell cycle alterations due to changes in Whi5 phosphorylation.
**4. Chromatin Immunoprecipitation (ChIP):**
The team used ChIP assays to examine how Whi5 binding to DNA was affected by its phosphorylation state. This involved cross-linking proteins to DNA, then immunoprecipitating Whi5-DNA complexes to analyze which genes Whi5 was regulating under different phosphorylation conditions. This method helped clarify how hypo- and hyper-phosphorylation modulated Whi5’s ability to inhibit or permit transcription of S phase-promoting genes.
**5. Biochemical Assays:**
Kinase assays were carried out to determine which kinases were responsible for phosphorylating Whi5 at different stages of G1. Additionally, phosphatase treatments were used in vitro to observe the dephosphorylation of Whi5, providing insight into the reversibility of these phosphorylation events and the potential regulatory roles of phosphatases in this process.
Through this array of methodological approaches, the study meticulously elucidated the complex regulation of the G1/S transition by Whi5 phosphorylation states. Each technique contributed to a holistic understanding of how modulation of these phosphorylation sites controls the cell cycle, paving the way for potential implications in more complex eukaryotic systems and offering insights that may lead to novel therapeutic strategies for cell proliferation-related diseases.
Continuing from the rigorous methodologies employed, the key findings of the study brought significant insights into the regulation of the cell cycle by phosphorylation dynamics of Whi5.
**1. Identification of Critical Phosphorylation Sites:**
The mass spectrometry analysis revealed 19 phosphorylation sites on Whi5. Particularly, seven of these sites were critical during the hypo-phosphorylated state in the early G1 phase. Mutations at these sites, converting serine or threonine to alanine, led to Whi5 remaining in its hypo-phosphorylated form for an extended period, thereby delaying the transition to the S phase. This identified delay highlights how precise the phosphorylation timing must be for optimal cell cycle progression.
**2. Effects of Phosphorylation on Cellular Localization:**
Through fluorescence microscopy, it was observed that as hypo-phosphorylated Whi5 stays predominantly in the nucleus during early G1, it effectively inhibits the transcription of S phase genes by binding to their promoters. Upon hyper-phosphorylation, Whi5 rapidly translocates to the cytoplasm, which correlates with the activation of these genes and progression into the S phase. These dynamic shifts underscore the importance of spatial regulation of Whi5 and its exit from the nucleus, which is pivotal for cell cycle continuation.
**3. Role of Priming Phosphorylation Sites:**
The study unearthed the concept of priming phosphorylation sites which are essential for the rapid phase of hyper-phosphorylation. These priming sites, once modified, facilitate subsequent phosphorylations at other sites. This sequential phosphorylation process is critical for the timely exit of Whi5 from the nucleus. These results show a sophisticated layer of regulation where initial modifications set the stage for more complex downstream effects, essential for precise cell cycle control.
**4. Impact on Cell Size and Growth:**
Further exploring the consequences of phosphorylation modifications, the findings indicated that changes in the phosphorylation pattern of Whi5 not only impact the timing of cell cycle transitions but also affect overall cell size and growth rate. Cells with hypo-phosphorylation-mimicking mutations tended to grow larger before transitioning to the S phase, implying that the regulation of Whi5 is fundamental to integrating growth signals with cycle progression cues.
**5. Biochemical Interactions and Kinase Activity:**
Kinase assays pinpointed specific kinases responsible for the phosphorylation at different stages, with particular emphasis on the Cdc28 kinase, well known for its role in cell cycle regulation. This interaction was not unidirectional; the phosphorylated state of Whi5 influenced the activity of various kinases, suggesting a feedback mechanism where Whi5 not only responds to but also modulates kinase activity.
These findings enrich the understanding of cell cycle regulation through a new lens, emphasizing the complexity and precision of phosphorylation patterns in controlling key transition points. By manipulating these phosphorylation sites, the researchers demonstrated a direct link between molecular modifications and broader cellular outcomes, such as growth, size regulation, and division timing. This study thus not only advances our knowledge of cell biology in yeast but also provides a framework that can be extrapolated to more complex eukaryotic systems, offering insights with potential therapeutic benefits against diseases characterized by cell cycle dysregulation. This research stands as a model for exploring similar regulatory mechanisms in human cells, which could lead to innovative approaches for managing proliferative diseases like cancer.
As the study by Jordan Xiao and colleagues unfolds the complex layers of the cell cycle regulated through phosphorylation dynamics of Whi5, it paves the way for more extensive research in this domain. The meticulous detail with which phosphorylation sites have been mapped and their functional implications studied in yeast serves as a robust foundation for exploring similar mechanisms in higher organisms, particularly humans. The parallels drawn between Whi5 in yeast and the retinoblastoma (Rb) protein in mammals suggest that deeper insights into such cell cycle regulators could lead to breakthroughs in understanding how the cell cycle is dysregulated in cancer and other proliferative diseases.
### Future Directions:
**1. Comparative Studies in Higher Eukaryotes:**
Building on the findings from this yeast model, future research should focus on comparative studies in more complex eukaryotes. This includes investigating analogous proteins to Whi5, such as the Rb protein, to determine if similar phosphorylation patterns and mechanisms dictate cell cycle progression in mammalian systems. Such comparative analyses may reveal evolutionary conserved strategies in cell cycle regulation that are pivotal for developing cross-species therapeutic targets.
**2. Integration of Systems Biology Approaches:**
Employing systems biology to integrate data from genomics, proteomics, and metabolomics could offer a holistic view of the cell cycle regulatory networks. This multidimensional approach might help in understanding how different cellular signals and environmental conditions influence phosphorylation events, thereby affecting cell cycle progression.
**3. Development of Targeted Therapies:**
With a comprehensive understanding of essential phosphorylation sites and their roles in cell cycle transitions, pharmaceutical researchers can design inhibitors that mimic or block these phosphorylation interactions. Such targeted therapies could be crucial in treating cancers where cell cycle regulation is disrupted. For instance, developing kinase inhibitors that specifically target the enzymes responsible for the hyper-phosphorylation of cell cycle inhibitors could halt the progression of cancer cells.
**4. Advanced Imaging and Real-Time Tracking:**
Technological advancements in live-cell imaging and real-time tracking of molecular interactions within cells will refine our understanding of the dynamic processes governing cell cycle phases. Enhanced imaging techniques could allow researchers to observe the immediate effects of experimental manipulations on phosphorylation states and cellular behavior.
**5. Public Health and Preventive Medicine Applications:**
From a public health perspective, understanding cell cycle regulation at a molecular level could facilitate the development of predictive diagnostics that assess risk levels for cell cycle-related diseases, including various cancers. This could lead to preventive interventions tailored to the molecular phenotypes observed in individuals.
### Final Thoughts:
The intricate dance of phosphorylation within a cell as it progresses through its cycle highlights not just the complexity of biological systems but the exquisite precision needed to maintain health. The work by Xiao and team not only advances our fundamental understanding of cell biology but also frames a narrative of potential translational research where the knowledge gained in yeast cells might one day inform human medical treatments. As we move forward, the integration of interdisciplinary research approaches will undoubtedly be pivotal in harnessing the full therapeutic potential of cell cycle regulation. This research exemplifies the profound impact of fundamental science on potential future applications in medicine and health, marking just the beginning of a promising avenue of exploratory and applied research in cell cycle dynamics.